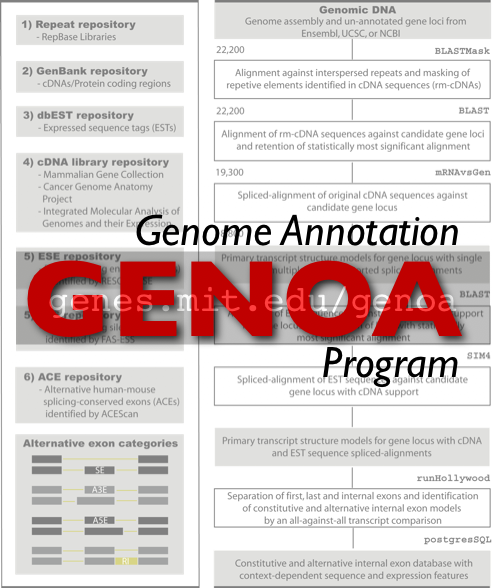
D. Holste, C.B. Burge, et al. The making of mRNAs: computationally dissecting the alternative splicing of precursors by using the Hollywood database. In preparation. World-wide web Supplementary material
D. Holste, G.Huo, V.Tung, and C.B. Burge. Hollywood: a comparative genomics relational database of alternative splicing. In preparation. World-wide web Supplementary material
E. van Nostrand, D. Holste, Burge CB. Orthology-based characterization of human intron retention. In preparation. World-wide web
G.W. Yeo, E. van Nostrand, D. Holste, Poggio T, Burge CB. Identification and analysis of alternative splicing events conserved in human and mouse. Proc Natl Acad Sci USA 102(8):2850 (2005). World-wide web Supplementary material
G. Yeo*, D. Holste*, G. Kreimann, and C.B. Burge. Variation in alternative splicing across human tissues. Genome Biol 5(10):R74 (2004). World-wide web Supplementary material
W.G. Fairbrother, D.Holste, C.B.Burge, and P.A. Sharp. Single nucleotide polymorphism-based validation of exonic splicing enhancers. PLoS Biol 2(9):E268 (2004). World-wide web Supplementary material